DECODR 2014: Data Processing Notes, sta.csv version
DECODR 2014 is SIO-CalCOFI's data processing suite which processes and merges the bottle & CTD data collected on each CalCOFI station. DECODR combines CTD sensor measurements with the corresponding depth bottle data into station data files (75+ sta.csvs). Station and cast information, such as weather, are stored in a single casts file (casts.csv). The casts & sta.csv formats, adopted in 2011, replaces the legacy data processing file formats based on 128-col computer punch cards: 00, 20, 22, W3, stacst, wea & IEH. The SIO-CalCOFI hydrographic database, Cruise Data Reports, and IEH archive data files are products generated by DECODR from casts.csv and sta.csvs.
CTD Data
CTD data processing is done using Seasoft, Seabird's data processing software (please refer to the CTD Data Processing Manual for specifics). DECODR 2014 will import 1m (or 4-second) binavg matching-depth CTD data into the sta.csvs. Early in data processing, the importing of CTD data is very useful for bottle vs CTD data quality control. The early cross-checking of bottle and CTD data expedites data quality assessment for both data types, discreet seawater samples and electronic sensor measurements. When compared on the cruise, early detection of bottle mistrip issues or sensor failure is possible.
Once bottle data are final, these data are used to bottle-correct the CTD sensor data. The preliminary CTD data in the sta.csvs, used during point-checking, are replaced by final, bottle-corrected CTD sensor data and hybrid (bottle and CTD) data products are published. Standard depth data, which may be interpolated when necessary, are often replaced with bottle-corrected CTD sensor measurements (temperature, salinity, and oxygen).
Seawater Sample Processing
 Each sample assay generates a data file for each station - some assays may combine multiple stations in a run into one file. These data files reside in specific directories for each data type. Each data processing module will parse the runs in their respective subdirectories and build a station list to process.
Each sample assay generates a data file for each station - some assays may combine multiple stations in a run into one file. These data files reside in specific directories for each data type. Each data processing module will parse the runs in their respective subdirectories and build a station list to process.
Processing all of one sample type is performed in batch, in similar fashion, for each datatype: salts, oxygens, chlorophylls, nutrients, and primary productivity samples. Stations may be processed in smaller groups or individually by checking or unchecking the station(s) to process. Each data processing module generates an output file of calculated measurements for examination and assessment of data issues & quality.
Each processing module ingests data files generated during the analysis of each seawater sample. As needed, the data files are manually 'cleaned-up' - edited and corrected - using a text editor such as Ultraedit then reprocessed. The output from edited files are once again checked for data quality and the process repeated until the data files are consider 'point-check ready'. Then the data from each assay are merged into station files (sta.csvs) by rerunning each DECODR modules with the 'Update CSVs' option checked. This populates the casts.csv with cast information and sta.csvs with bottle data.
Datacodes are used to annotate the best data for publication. By default, bottle data are data coded for use to generate Data Reports, database csvs & IEH generation.
Casts.csv and sta.csvs are located in the CSV subdirectory. Each module run with generate backup files archived in the CSV\BAK subdir. Each backup is timestamped & flagged with the data update type.
DECODR Basics
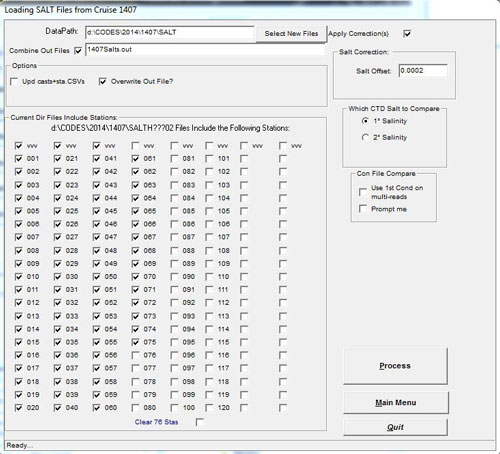
- Process Salts, Update Casts+Sta.csvs unchecked, will generate a bottle salt outfile with CTD 1° salt values from Portasal sample run files (Salt###). A salinity offset may be applied to selected stations if there is a noticable drift in the Sub Standard. The salinity offsets applied will be tabulated in the casts.csv (YYMMcasts.csv) for each station. Other options include 'Use 1st Cond on Multi-Reads' - when there are multiple reads, the 1st is often considered the best when subsequent conductivity readings drift upwards. Comparing the bottle salinity to primary or secondary CTD salts may help resolve drift issues. Once any problem with the salt files are corrected, re-run the salts with the Update Casts+Sta.csvs checkbox checked. The salt data output will be added to all the selected station csvs. This update may be performed on sta.csvs repeatedly but bottle data updates of any datatype should not be performed on terpled.csvs (t.csvs). SDC will be set to '0'.
- Process Oxygens, Update Casts+Sta.csvs unchecked, will generate an oxygen outfile from oxygen auto-titrator sample run files (YYMM###). This module has a Recalculate Normality option. Assess sample correctness using the outfile then manually correct individual oxygen files using a text editor. Once resolved, re-run the oxygens with Update Casts+Sta.csvs checked. This will add bottle oxygens to all the selected station files. This may be performed on sta.csvs at any stage of the data process except terpled files (t.csvs). ODC will be set to '0'.
- Process Chlorophylls, Update Casts+Sta.csvs unchecked, will generate a chlorophyll outfile from (FLog) chlorophyll fluorometric analysis files. This module has data processing options for Blank, Tau and Acid Ratio. Assess sample correctness using the outfile then manually correct individual chl files using a text editor. Once resolved, re-run the chlorophylls with theUpdate Casts+Sta.csvs checked. This will add bottle chlorophylls and phaeopigments to all the selected station files. This may be performed on sta.csvs at any stage of the data process except CSL-terpled files. CPDC will be set to '0'.
- Process Nutrients, Update Casts+Sta.csvs unchecked, will generate a combined nutrient outfile from QuAAtro data txt files (YYMM###.txt or YYMM###R#.txt). Adjust the data processing parameters on the right, particularly the MDL ranges and options. Combined station runs need to be delimited by a "STA###" line separating one station from another. Assess data quality using the outfile then manually correct the appropriate run.txt file using a text editor. Once resolved, re-run the nutrients with Update Casts+Sta.csvs checked. This will add core nutrients and ammonia to all the selected station csvs. This may be performed on sta.csvs at any stage fo the data process except CSL-terpled files. NDC & NHDC will be set to '0'.
- Process Prodos, Update Casts+Sta.csvs unchecked, will generate a primary productivity outfile. Processing prodos requires the correct importing of the scintillation counter file using DECODR module Import Scint. Counts to generate YYMMscin.csv. This module has fields for Specific Activity and Blank coefficients, and different scint data filenames. Assess sample matching correctness using the outfile then manually corrected individual prd files or scint asc file using a text editor. Once resolved, re-run the prodos with Update Casts+Sta.csvs checked. This will add 14C uptake data to all the selected prodo station csvs. This may be performed on sta.csvs at any stage of the data process except CSL-terpled files. C14DC will be set to '0'.
-
Import CTD .btl files. Run DECODR's [CTD Data to CSV] module and select .btl files. Btl files should be generated by the primary CTD data processor & include the CTD sensor & derived data fields listed below. The default settings should be fine for the update. CTD preliminary asc files or bottle-corrected CTD.csvs will be used for subsequent CTD data updates.
- To generate .btl files - run Seasoft's Datcnv module and select 'create bottle (.ROS) file' under Data Setup; Scan range offset = -4, Scan range duration = 4. Be sure to select all data needed to derive the CTD data columns. This will generate .ros files for each cast.
-
Run Seasoft's Bottle Summary with the following Averaged CTD sensor data & Derived data.
- Averaged variables: Scan Count, Julian Days, Depth salt water m, Pressure digiquartz db, Temp 1 ITS-90 degC, Temp 2 ITS-90 degC, V0 - V7, Beam Attenuation Seatech, Beam Transmission Seatech, Fluorescence Seapoint, pH, Lat deg, Lon deg
- Derived Variables: Sal 1 PSU, Sal 2 PSU, O2 SBE 43 ml/L, O2 SBE 43 2 ml/L, Density sigma-theta Kg/m^3, Density 2 sigma-theta Kg/m^3, Oxygen SBE 42 % saturation, Oxygen SBE 42 2 % saturation, Potential Temp ITS-90 deg C, Potential Temp 2 ITS-90 deg C
- Be sure the 'Output min/max values for averaged variables' box is checked along with 'Apply Tau correction'. This will generate the .btl files which can be imported into the sta.csvs.
-
Import CTD .csv files. Run DECODR's [CTD Data to CSV] module and select .csv files. CTD.csv files should be generated by the primary CTD data processor. These are the bottle-corrected CTD + bottle data csvs generated by BtlVsCTD step 2. The default data options for the different data types should be good unless you are comparing legacy data to sta.csv data, then select 'do not update the primary & secondary temperatures'. This will improve the agreement since 4sec .btl data are considered closest to bottle data. All derived bottle data use these temperature to generate the calculated values such as SVA.
- For the first update, select Update Bottle Depths Only. This will update the CTD data in the sta.csvs - use the selection matrix to pick the desired data such as Corrected Primary Salinity & Sta Corr Oxygen.
- Then select Add CTD StndLvls, Terple Bottle Data is automatically included. If you want the SDC & ODC codes to be set as '1' for CSLs, select those options in the Data Codes. When standard levels are inserted, new sta.csvs are generated, preserving the observed data sta.csvs for further updates. These files are named YYMM###t.csv.
- Point-check the CSL-terpled YYMM###t.csvs but correct YYMM###.csvs. If CSL-terpled csvs are corrected, interpolated bottle values will not be re-interpolated. This may be a future feature but for now all corrections should be applied to observed data files only.
- Update sta.csvs then generate final CSL-terpled t.csvs by re-running DECODR's [CTD Data to csv] module with final sta.csvs and Add CSLs StndLvls & Terple Bottle Data boxes checked.
-
Generate data products
- Cruise Data Report using Data Rpt from CSVs module
- Cruise Primary Productivity Report using ProdoRpt from CSVs module
- Horizontal & Vertical Section Plots using Horizontals & Verticals Contouring modules
- Database Bottle & Cast csvs using Sta.csvs>bottledb.csv & Casts.csv>castdb.csv modules
- IEH using Make IEH from CSVs module
General Notes:
DECODR uses C:\DECODR\decodr.cfg to configure the program. It is mandatory for it to be in that specific dir. The executable may be run from any location but the cfg file has to be local. The default datapath is the root data dir YYMM. The SUMMARY dir datapath is set by changing the '0' to '1' in cfg line 9. Or by selecting the SUMMARY box in DECODR's main window (upper right).
decodr.cfg explained
| Settings | Description |
| 1411 1407 1404 1402 1311 | Current Cruises being processed |
| 1407 | Last cruise picked |
| C:\CODES\2014\1407\ | Default datapath; SUMMARY directory is retired* |
| C:\CODES\2014\1407\ | Datapath of last run |
| C:\Decodr\ | Decodr default exe path |
| 100 | Max salinity samples per data file |
| JRW DMW JLW DNF MGS | Initials of Primary Processors |
| 22.5 1.80 45.0 0.75 3.00 | Nutrient Standard Concentrations - format is important! xx.x x.xx xx.x x.xx #.## |
| 0* | Summary dir? 0=No, 1=Yes (Retired in 2011) |

